- HOME
- News & Events
- Publications
- 【Publications】Single-cell atlas of lineage states, tumor microenvironment and subtype-specific expre...
Publications
【Publications】Single-cell atlas of lineage states, tumor microenvironment and subtype-specific expression programs in gastric cancer
October 27 2021
Lab: Takatsugu Ishimoto
Paper information
Title:
Single-cell atlas of lineage states, tumor microenvironment and subtype-specific expression programs in gastric cancer
Kumar V, Ramnarayanan K, Sundar R, Padmanbahan N, Srivastava S, Koiwa M, Yasuda T, Koh V, Huang KK, Tay ST, Ho SWT, Tan ALK, Ishimoto T, Kim G, Shabbir A, Chen Q, Biyan Z, Xu S, Lam KP, Lum HYJ, Teh M, Yong WP, So JB, Tan P*.
(*corresponding authors)
Cancer Discov. 2021 Oct 12:candisc.0683.2021.
doi:10.1158/2159-8290.CD-21-0683.
Summary
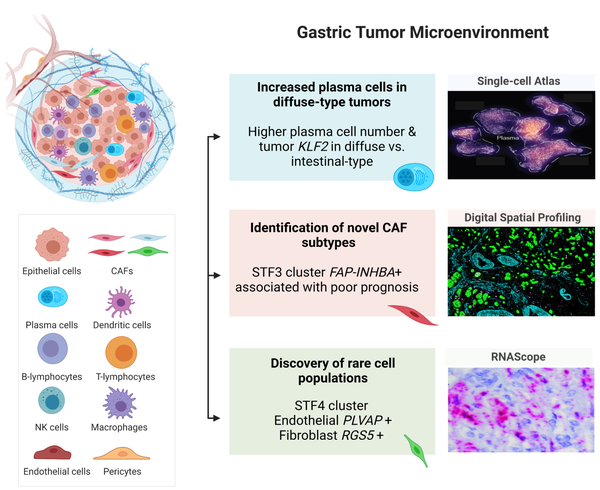
Gastric cancer (GC) heterogeneity represents a barrier to disease management. We generated a comprehensive single-cell atlas of GC (>200,000 cells) comprising 48 samples from 31 patients across clinical stages and histological subtypes. We identified 34 distinct cell-lineage states including novel rare cell populations. Many lineage states exhibited distinct cancer-associated expression profiles, individually contributing to a combined tumor-wide molecular collage. We observed increased plasma cell proportions in diffuse-type tumors associated with epithelial-resident KLF2, and stage-wise accrual of cancer-associated fibroblast sub-populations marked by high INHBA and FAP co-expression. Single-cell comparisons between patient-derived organoids (PDOs) and primary tumors highlighted inter- and intra-lineage similarities and differences, demarcating molecular boundaries of PDOs as experimental models. We complemented these findings by spatial transcriptomics, orthogonal validation in independent bulk RNA-seq cohorts, and functional demonstration using in vitro and in vivo models. Our results provide a high-resolution molecular resource of intra- and inter-patient lineage-states across distinct GC subtypes.